Overview

Since the earliest invention of telescopes, microscopes, and eyeglasses, imaging systems have been designed to help humans visualize the world around us – big and small, near and far. These imaging systems collect the light reflected or emitted from an object and focus it onto our eyes or a camera. The design of these systems dictates that their images only contain two-dimensional (2D) information about an object and that their 2D images are blurred if the object is out of focus. We build imaging systems with new capabilities that surpass these shortcomings.
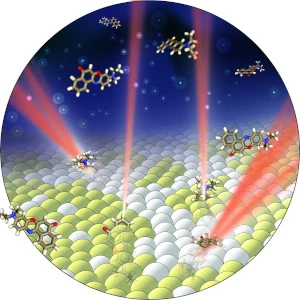
Super-resolution is a key feature of many of our imaging systems–the ability to overcome the resolution limit of wave physics, called the diffraction limit, in order to visualize the nanoscale world. Single-molecule imaging is also a central theme of our research–enabling our technology to see individual molecules as they drive biological and chemical dynamics at the nanoscale.
Our research

Video vignettes
Learn more
- Background on super-resolution imaging
- Our software - GitHub repository
- Data supporting our publications - OSF



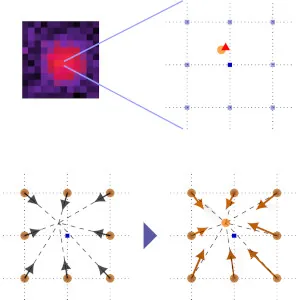
 How can we design
new dipole- and point-spread functions for
6D (3D position & 3D orientation) imaging?
How can we design
new dipole- and point-spread functions for
6D (3D position & 3D orientation) imaging?


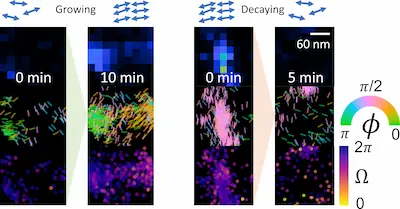
 Any image analysis algorithm (both physics-based methods and
data-driven deep learning techniques) uses an internal model of
the imaging system to estimate the parameters of interest (e.g.,
the position of a single molecule). How can we detect if the model
is incorrect for some or all of the observed data
without knowing the ground truth?
Any image analysis algorithm (both physics-based methods and
data-driven deep learning techniques) uses an internal model of
the imaging system to estimate the parameters of interest (e.g.,
the position of a single molecule). How can we detect if the model
is incorrect for some or all of the observed data
without knowing the ground truth?